Welcome to EURenOmics!
Although the EURenOmics project officially ended on September 30th, 2017, the group continues to be devoted to improving the lives of patients affected by rare kidney diseases. In line with the objectives of the International Rare Disease Research Consortium (IRDiRC), we aim to develop novel tools that will allow to make more accurate diagnoses, predict the disease course and the efficacy of available treatments, and help developing new and better therapies for rare kidney diseases. A summary of the main achievements in year 5 can be found here and a summary of the 5-year project duration can be found here.
The EURenOmics project received funding as part of the European Community’s commitment to the IRDiRC initiative. In this quest we joined forces with our FP7 partner project Neuromics (for neuromuscular disorders) and the RD-CONNECT infrastructure platform.
The research efforts of EURenOmics were focused on the following 5 groups of kidney diseases:
- Steroid resistant nephrotic syndrome (WP2)
- Membranous nephropathy (WP3)
- Tubulopathies (WP4)
- Complement disorders such a haemolytic uraemic syndrome (WP5)
- Congenital kidney malformations (WP6)
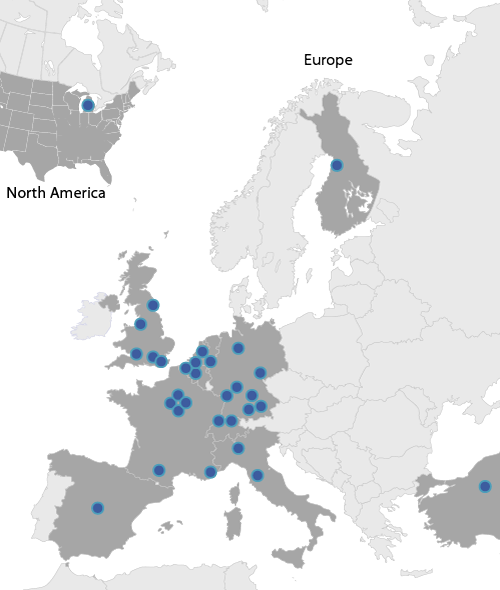
Our Consortium comprised 17 European academic institutions, 9 SMEs and 1 academic partner from the United States. We have access to the largest rare renal disease cohorts assembled to date (collectively >12,000 patients) with detailed clinical information and comprehensive biorepositories containing DNA, blood, urine, amniotic fluid and kidney tissue.
In EURenOmics we utilized a wide array of high-throughput technologies to find new genes causing or predisposing to kidney diseases, characterize molecular signatures uniique to individual disease entities, identify prognostic biomarkers, and screen for potential drug candidates. These technologies include next generation exome and whole-genome sequencing, ChiP-sequencing, tissue transcriptome and epitope profiling, and miRNome, proteome and metabolome screening in different body fluids.
The resulting data are being combined in a systems biology approach with high-resolution clinical phenotyping and findings obtained with a large array of established and novel in vitro, ex vivo and in vivo disease models to identify disease-associated genetic variants, disease-defining molecular signatures, and potential targets for therapeutic intervention.
We are deeply grateful for the trust and support we received from our professional societies, i.e. the European Society for Pediatric Nephrology, the International Pediatric Nephrology Association, and the European Renal Association (ERA-EDTA) with its Work Group for Inherited Kidney Diseases (WGIKD), as well as from the numerous patient organizations and advocacy groups in Europe and around the globe that are following our efforts with much attention and sympathy.