Genomic research
The Consortium encompasses a group of nephrogenetic researchers with a superb track record in identifying and characterizing disease causing genes. Their concerted search for the remaining renal disease genes, with shared access to cohorts of familial cases via the registries, will be a particular strength of the EURenOmics project. Besides the search for new genes causing monogenic disorders, the group will jointly tackle the exploration of complex genetic disease transmission in rare kidney disorders. The next-generation sequencing core facilities are uniformly equipped with latest state-of-the-art technologies. The researchers will share their expertise in applying filtering strategies and comparisons with reference exome data. Genome-wide ChipSeq analysis and the human fetal to adult kidney tissue expression profiles available through the Consortium will deliver important additional information at the transcriptional level. The Bioinformatics Core will provide comprehensive assistance in candidate gene definition by providing a unique set of data mining tools.
Apart from identifying new disease genes, the group will be developing efficient multiplex PCRs (MASTR assays) for rapid disease-group specific targeted massive parallel sequencing. This important methodological work will be carried out in cooperation with the SME partner Multiplicom Inc. and external collaborator Karin Dahan (University of Louvain), who has extensive recent expertise with this novel technology.
Molecular Signature and Biomarker Research
In the field of -omics profiling the Consortium contains several SME’s, each of which has demonstrated leadership in the scientific development and practical application of molecular signatures. The partner mosaiques diagnostics has been leading the field of urinary proteomics over the past decade, with more than 100 publications. The company’s proprietary technological platform has been approved by EMA and FDA for use in pharmaceutical studies. Mosaiques maintains a database containing >20.000 proteomic datasets from healthy controls and patients with a variety of relevant diseases, which will be of tremendous value in identifying signature patterns and/or individual biomarkers specific to the phenotypes of the rare renal diseases of interest.
Figure 1: Pattern-comparison healthy versus diseased
Likewise, Metabometrix Ltd. is an international leader in the field of metabonomics. The partner uses advanced spectroscopic and analytical techniques and a range of proprietary statistical algorithms to create novel metabolic fingerprints defining a particular disease condition, the state of disease activity or the response to pharmacological treatment. Metabometrix and University College London have been pioneering the use of metabonomics to derive molecular signature in rare kidney disease; their joint experience will be of great value in the further exploration of the clinical usefulness of this approach in EURenOmics. Comprehensive Biomarker Center (CBC) is a young but already well-established company focusing on the application of mrRNAomic profiling in various body fluids to establish diagnostic signatures in human disease conditions. CBC has been involved in the recent definition of the human serum miRNAome. One of CBC’s key areas of expertise is the development of highly efficient miRNA extraction methods; the company will contribute to the project by establishing miRNA extraction from urinary exosomes in partnership with Radboud University Nijmegen Medical Center.
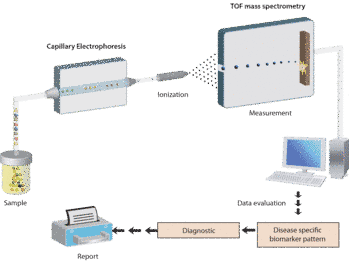
Figure 2: Proteomic analysis of urinary biomarkers.