WP02 - Steroid Resistance Nephrotic Syndrome
Objectives
- Identification and functional characterization of new disease-causing and modifier genes underlying SRNS
- Development and validation of a new disease ontology for SRNS by integration of phenotypic and genetic infor-mation with specific molecular signatures derived from tissue and urine analysis
- Development and implementation of rapid diagnostic tools and biomarkers
- Develop novel molecular therapies for genetic SRNS
Workpackage Description
Childhood nephrotic syndrome is a severe clinical condition characterized by a loss of glomerular permselectivity with massive urinary protein loss and consecutive alterations of lipid metabolism, haemostasis and endocrine systems. The etiologically heterogeneous condition is characterized by dysfunction of the podocytes, the visceral epithelial cells which form the glomerular filtration barrier. Two fundamental mechanisms are believed to cause podocyte dysfunction in childhood nephrotic syndrome: (i) an abnormal response of the innate and/or inducible immune system to exogenous stimuli (e.g. infectious pathogens, toxins, allergens), and (ii) defective expression of podocyte-specific proteins resulting from inherited genetic abnormalities. Whereas the majority of cases are caused by an acute dysregulation of the immune system readily responsive to glucocorticoids, 20% suffer from steroid-resistant nephrotic syndrome (SRNS), a much more challenging disease entity frequently progressing to ESRD. Up to 50% of SRNS patients achieve complete or partial remission upon intensified immunosuppressive treatment. The remaining “multi-drug resistant” cases are at high risk of progressing to ESRD, however with a highly variable, poorly predictable individual rate of disease progression. Many of these cases are assumed to be due to genetic causes. A number of genes essential for podocyte development, structure and function have been identified and several additional loci have been linked to familial SRNS, supporting marked genetic heterogeneity. Altogether, abnormalities in the disease-associated genes identified to date explain only about 25% of familial and 10% of sporadic cases, and display large phenotypic variability. The preferential onset of genetic cases in the first decade of life and the 20% post-transplant SRNS recurrence rate suggest that a major fraction of SRNS cases will not be explained by abnormalities in podocyte-specific genes.
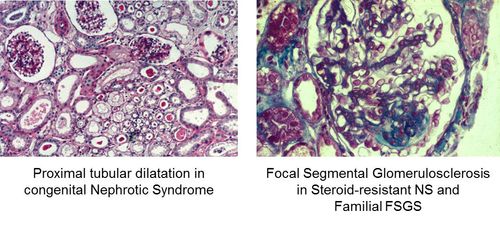
- Figure1: Staining of tissue samples from patients with hereditary nephrotic syndrome showing charactistic cellular changes.
Identification and characterization of novel disease causing and modifying genes underlying SRNS
This will be achieved through exome sequencing of 100 familial (either autosomal dominant or autosomal resistant) SRNS cases without mutations in known genes combined with genome-wide linkage analysis to identify novel SRNS genes and whole-genome sequencing of 100 selected sporadic cases (multidrug resistant patients below 5 years of age, without mutations in the known SRNS genes) to catalogue all rare variants in the coding sequence and regulatory sequences (cf below) of all podocyte genes to assess the true frequency of monogenic and oligogenic cases. Genomic key regulatory elements will be identified by ChIP-Seq analysis via the identification of the genome-wide distribution of active epigenetic marks (H3Ac, H3K4me3 etc.) in podocytes and the binding sites of specific transcription factors (WT1/Lmx1B/mafB etc.), and followed up with comprehensive bioinformatic screening
The normal and mutated gene products of the newly discovered disease associated genes will be characterized by studies of their expression pattern and subcellular localization as well as by functional studies in podocyte cell culture and animal models (zebrafish morpholino knock-down or transgenic, KO mice if available).
Deep phenotyping towards new disease ontology for steroid resistant nephrotic syndrome
- Definition of common data elements describing clinical phenotypes (e.g. age at onset, extrarenal symptoms, histopathological findings, pharmacological treatment responses, evolution of proteinuria and GFR, post-transplant disease recurrence); retro- and prospective phenotyping of all available SRNS/FSGS cohorts (paediatric: PodoNet, RADAR (UK); adult: Nijmegen and Bristol FSGS cohorts; paediatric and adult: Paris cohort and Neptune study (US)).
- Genotyping of all sporadic cases in PodoNet cohort for all known SRNS genes using novel multiplex-PCR/NGS kit as described below.
- Comparative urine peptidome and miRNAome profiling in 50 defined genetic, 50 multidrug-resistant non-genetic, 60 calcineurin-inhibitor (CNI)-responsive SRNS cases from PodoNet cohort (30 in remission, 30 during relapse) and 50 healthy controls matched for age and gender).
- Standardized prospective monitoring and sample collection (serum, urine, optional biopsy specimen) of 40 paediatric cases with defined genetic SRNS and 40 CNI-responsive and 40 multidrug-resistant cases (50% paediatric / 50% adult). Analysis of urine metabolome; urine exosome preparation for proteome and miRNAome profiling; glomerular transcriptome and miRNAome analysis in biopsy tissue processed along the ERCB protocol. Cross-validation of differentially expressed transcripts and miRNA on ERCB, NEPTUNE and RaDaR data and laser-micro dissected archival samples with long-term follow-up; comparison with results from adult membranous NP patients studied with identical protocols.
Development of diagnostic tools and prognostic biomarkers
- Development of a gene diagnostic tool using a combination of multiplex PCR and high throughput sequencing. This test will screen simultaneously 23 genes known to be involved in non syndromic forms of SRNS. The kit will be complemented by the new disease-associated genes identified in the course of the project to yield a commercializable diagnostic kit.
- Biomarkers of disease progression and post-transplant recurrence will be sought by comparative analysis of urinary peptidome, miRNAome and tissue mRNA expression patterns in prospectively followed cases, validated by analysis of archived cross-sectional samples with retrospective follow-up and longitudinal cross-validation cohorts (Neptune, RADAR) and compared with other progressive kidney diseases studied by the consortium.
High-throughput compound screening in cell models of genetic SRNS
High-throughput screening of compound libraries for active small molecular compounds restoring intracellular trafficking of ER-retained mutated podocyte membrane proteins (nephrin, podocin) will be performed using in vitro bio-assays recently set-up by partner 2a and the Biophenics platform (Curie Institute, Paris). Cell lines stably expressing wild-type and mutant, untagged or GFP-tagged podocin and nephrin have been established and used to automatize all the compound library screening steps, including fluorescence imaging in microplates and computational phenotyping of digital images. The Prestwick (1,200 molecules, 90% being marketed drugs and 10% bioactive alkaloids or related substances, thus presenting the greatest possible degree of drug-likelihood) and the Chembridge DIVERSet (10,000 to 50,000 drug-like, small molecules, with maximum pharmacophore diversity) libraries will be screened at the Biophenics platform. When compounds that target podocin and nephrin to the plasma membrane are identified, the functionality of the “rescued” proteins will be tested in vitro. in vivo testing of candidate compounds (provided their safety has been proven in animals) will be performed in a recently established inducible knock-in mouse model of the podocin mutation tested above. Studies will include dose finding series; endpoints will be proteinuria, global kidney function and renal histopathology at 4 weeks. Extensive screening for off-target (extrarenal) effects will be performed.
High-throughput compound screening in Zebrafish models
Zebrafish models including a nphs2 knock-out line (using zinc finger nuclease technology) and WT and R138Q podocin-tagged transgenic lines using the Gal4/UAS technology (a Nphs2::gal4 line is already available at our collaborator lab) will be generated in collaboration with I.Drummond, Boston. Given the phenotype of the Nphs2 knock-down by morpholino, a severe phenotype with cardiac edema, proteinuria and tubulocystic dilation is expected. The R138Q-podocin-GFP transgenic line is not expected to present any phenotype given the presence of the normal zebrafish alleles, but the mislocalized GFP-tagged mutant allele could be easily detectable. These models will then be used for high-throughput drug screening to identify molecules rescuing the phenotype (null allele model) or retargeting the mutant podocin to the plasma membrane (transgenic model). Fully automated screening system using excretion of fluorescent protein and/or pattern recognition (edema, tubulocystic).
WP Leader
Prof. Corinne Antignac (Deputy: Franz Schaefer)
INSERM U983
Participating Partners
Heidelberg University Medical Center
INSERM U983
INSERM UMRS 970
University of Zurich
Istituto di Recerche Farmacologiche Marion Negri
University of Michigan
Hacettepe University
University of Bristol
Radboud University Nijmengen Medical Center
Metabometrix Ltd.
Multiplicom Inc.
Comprehensive Biomarker Center
mosaiques diagnostics GmbH
Karlsruhe Institute of Technology